Omics
Two molecular markers associated with sucrose accumulation were used to validate genotyping techniques. Targeted sequencing techniques have shown promise for identifying markers in the sugarcane polyploid genome. Research progress.
Writers: Fernando S. Aguilar1, Alejandra Londono1, 2, Carolina Saavedra2, Hugo Arley Jaimes1, John Henry Trujillo1, Jershon López Gerena1 and John Jaime Riascos1

The alliance OMICS, In-silico Multiscale Optimization of Sustainable Agricultural Crops, is a cooperation program in the strategic focus of Food in Colombia, designed to provide agricultural solutions in terms of food security and productive sustainability through research and education in omic sciences.
In five years of activities (2017-2022), the results include the validation of analytical genotyping techniques, aimed at whole genome sequencing (WGS) and capable of reproducing SNP markers, that is, single nucleotide polymorphisms. . Genomic experimentation was carried out after sequencing 220 sugarcane genotypes (saccharum spp.) from the Cenicaña germplasm bank, and align the information to the complete genome of the variety CC 01-1940. With this information, associations were made and SNPs related to sucrose accumulation were selected.
What is expected from genotyping techniques?
- To accurately identify the position of a SNP-type marker in the sugarcane polyploid genome.
- Estimate the allelic dose of SNP markers at each position in the genome, that is, the number of reference alleles (those that are the same as those of the reference genome CC 01-1940) and alternative alleles (different from those of CC 01-1940 ).
- Increase the capacity to process a large number of genotypes (DNA samples) with speed and reliability.
- Reduce genotyping costs and gain efficiency in genomics studies.
Assist plant breeding in the production of high productivity genotypes.
Testing amplification and sequencing techniques
Cenicaña evaluated three genotyping techniques, two of them based on DNA amplification (KASP and ddPCR) and a third based on sequencing (Flex-seq).
- KASP (Kompetitive Allele Specific PCR) is based on an allele-specific competitive PCR that allows differentiating variations in a single nucleotide by changes in fluorescence levels, so that it is possible to assign the genotypic class to each individual (ie, homozygous or heterozygous). ) according to the count of the reference and alternative alleles.
- The ddPCR (Droplet Digital PCR) includes the partitioning of the DNA in multiple repetitions by means of the emulsion of water and oil for the formation of microdroplets (more than 18,000, each one with a volume close to 1 nanoliter). These droplets are analyzed by digital PCR to increase detection levels of SNP markers using specific fluorescent probes.
- Flex-seq® is a methodology based on sequencing directed to the adjacent regions of the SNP marker of interest (approximately 400 nucleotides). This methodology, developed by the company Rapid-Genomics, has the advantage of detecting the sequence of interest with greater precision and speed.
The three techniques made it possible to estimate the allelic dose of SNPs in each of the positions of the genome of the individuals evaluated.
Validating the reproducibility of allelic doses
The allelic doses in sugarcane can vary between 0 and 10 due to the polyploidy of the species, a much higher range than that of diploid organisms such as corn or rice, which vary between 0 and 2.
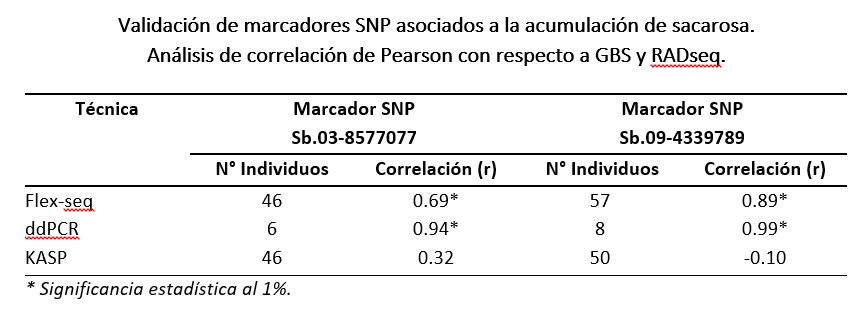
Thus, in order to verify the degree of reproducibility of SNP markers with the new techniques, the validation experiment was carried out to compare the allelic doses obtained with each one (KASP, ddPCR and Flex-seq) and those generated in previous years. by means of GBS (genotyping by sequencing) and RADSeq (restriction site associated DNA sequencing). For this purpose, the Pearson correlation coefficient was estimated, a measure of linear dependence between two quantitative random variables that is not affected by the scale of measurement of the variables.
Conclusions
- Flex-seq and ddPCR presented a correlation coefficient (r) greater than 0.69, which suggests that both techniques allow reproducing the results obtained by GBS and RADseq analysis.
- For KASP, a low correlation was observed with respect to GBS and RADseq, indicating low reproducibility of the technique for SNP sequences. In this case, none of the markers had fluorescence distinctive from a decaploid organism, so an assignment typical of diploid organisms was used. However, this assumption of diploidy resulted in a correlation coefficient lower than 0.32, indicative of low reproducibility of the technique.
- Flex-seq has a lower cost in data processing and has a high capacity to accurately identify the position of an SNP marker in the polyploid genome of sugarcane, which makes this technique ideal for marker validation. economically important molecules in sugarcane populations.
Finally, based on the results, it was decided to evaluate directed sequencing alternatives, that is, Flex-seq type techniques, seeking to reduce genotyping costs.
| SNP | Single Nucleotide Polymorphis | single nucleotide polymorphism | |
| wgs | Whole Genome Sequencing | Whole genome sequencing | (Neves et al., 2012) Neves, HHR, Carvalheiro, R., & Queiroz, SA (2012). A comparison of statistical methods for genomic selection in a mice population. BMC Genetics, 13. https://doi.org/10.1186/1471-2156-13-100 |
| GBS | Genotyping-By-Sequencing | Genotyping by sequencing | (Elshire et al., 2011) Elshire, RJ, Glaubitz, JC, Sun, Q., Poland, JA, Kawamoto, K., Buckler, ES, & Mitchell, SE (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE, 6(5), 1–10. https://doi.org/10.1371/journal.pone.0019379 |
| RADSeq | Restriction Site Associated DNA sequencing | Restriction site associated DNA sequencing | (Davey et al., 2011) Davey, JW, Hohenlohe, PA, Etter, PD, Boone, JQ, Catchen, JM, & Blaxter, ML (2011). Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Reviews Genetics, 12(7), 499–510. https://doi.org/10.1038/nrg3012 |
| KASP | Competitive Allele Specific PCR | Allele-specific competitive PCR | (He et al., 2014) He, C., Holme, J., & Anthony, J. (2014). SNP genotyping: the KASP assay. Methods in molecular biology (Clifton, NJ), 1145, 75–86. https://doi.org/10.1007/978-1-4939-0446-4_7 |
| ddPCR | Digital PCR droplet | Droplet digital PCR | Hindson et al., 2011 |
| flex-seq | sequencing directed to the adjacent regions of the SNP of interest |
____________________
1 Sugar Cane Research Center, Cenicaña
2 Pontifical Javeriana University, Cali















